Pupillometry Pipeline Guide
- This documentation will guide the researcher through the process of register, monitor & review pupillometry data.
Instructions to Set up Pupillometry Pipeline on a Rig Machine
- Ask your lab manager for helo to set up a camera in the rig.
- If needed, add these parameters in RigParameters.m
%% Pupilometry video parameters
video_parent_path = 'E:/VideoData'
video_ext = '.mj2'
video_acquisition_rate = 30
video_record = true
video_gain = 8
preview = true
- In Tranining GUI subject task selection menu, under
PupillometryVideoselect2. In RigParameters:
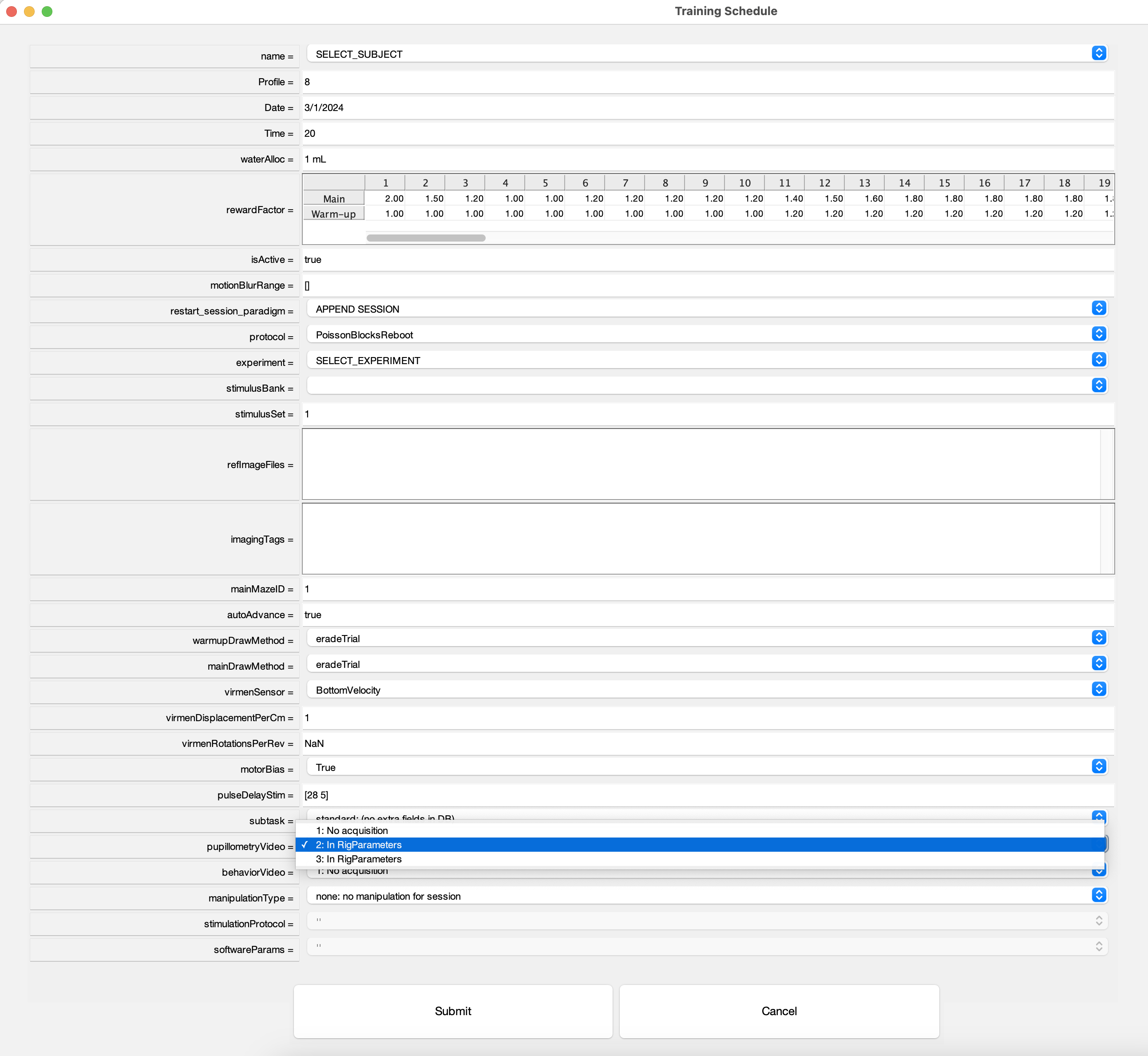
- In Experiment file add the following lines in
initializationcodeFunfunction (just aftervr = initializeGradedExperiment(vr);line):
% Start video acquisition
if ~isempty(vr.trainee.pupillometryVideo) && vr.trainee.pupillometryVideo ~= 1
vr = startVideoAcquisition(vr);
end
- In Experiment file add the following lines in
runtimeCodeFunfunction (just aftercatch err displayException(err);line):
% Stop video acquisition
if ~isempty(vr.trainee.pupillometryVideo) && vr.trainee.pupillometryVideo ~= 1
vr = stopVideoAcquisition(vr);
end
- In Experiment file add the following lines in
terminationCodeFunfunction (the very first line):
% Stop video acquisition
if ~isempty(vr.trainee.pupillometryVideo) && vr.trainee.pupillometryVideo ~= 1
vr = stopVideoAcquisition(vr);
end
Pupillometry backup videos task schedule
- On Windows type "Task Scheduler"
- Open Task Scheduler "App"
- On right hand side menu, click on "Create Task" Action
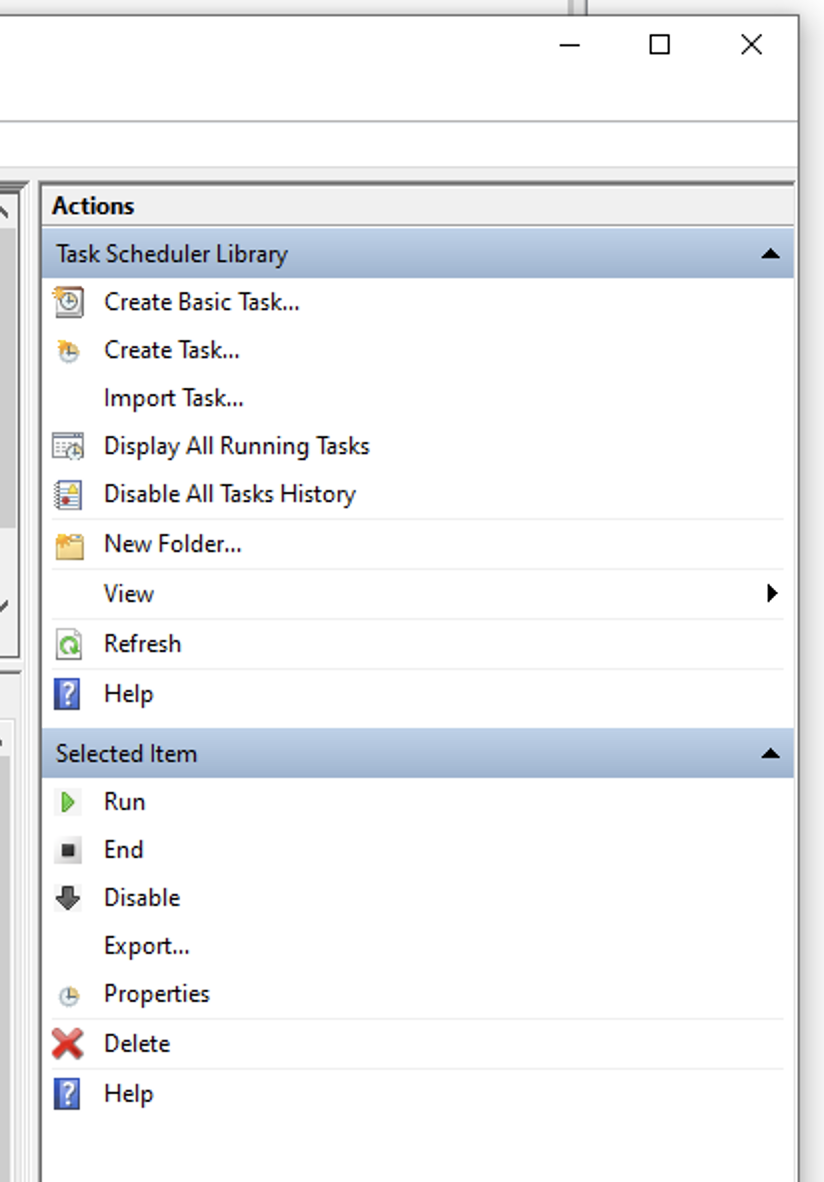
- Name new task as video_backup
- Add a trigger to run task daily at 11:30 pm
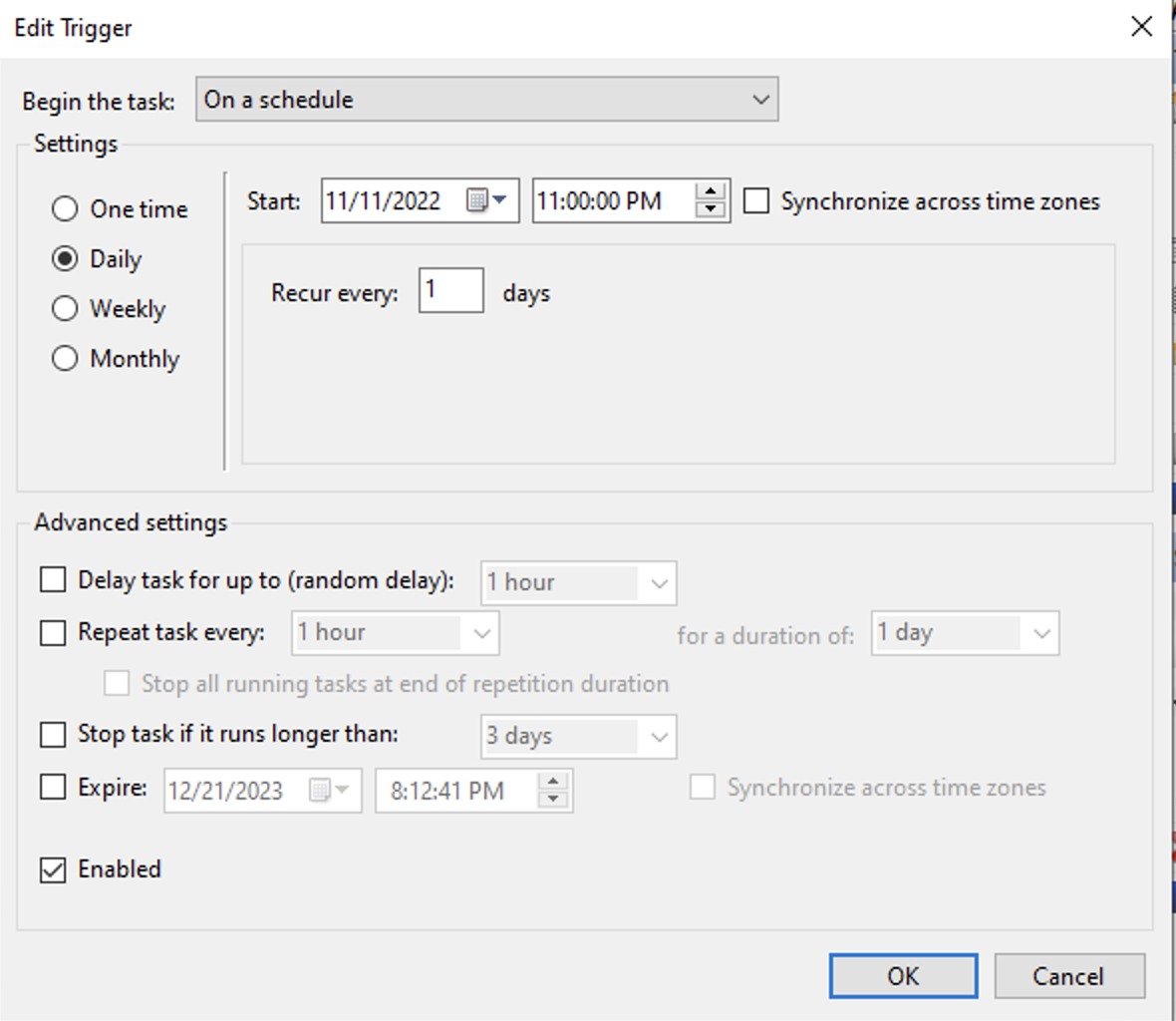
- Add an action: add this line to the Program/script edit:
C:\Experiments\U19-pipeline-matlab\scripts\cmd_copy_video_files - Hit OK button
Pupillometry DB and data organization
Raw data (video files) are located in:
\\cup.pni.princeton. edu\braininit\Data\Raw\video_pupillometry\(userid)\(subjectfullname)\(session_date)_g(session_number)\(video_file)Processed data (h5 files) are located in:
\\cup.pni.princeton. edu\braininit\Data\Processed\video_pupillometry\(userid)\(subjectfullname)\(session_date)_g(session_number)\(h5_file)Video models are located in:
\\cup.pni.princeton. edu\braininit\Data\Raw\video_models\(video_model_directory)Check DB Organization (pupillometry) for a description for each pupillometry table.
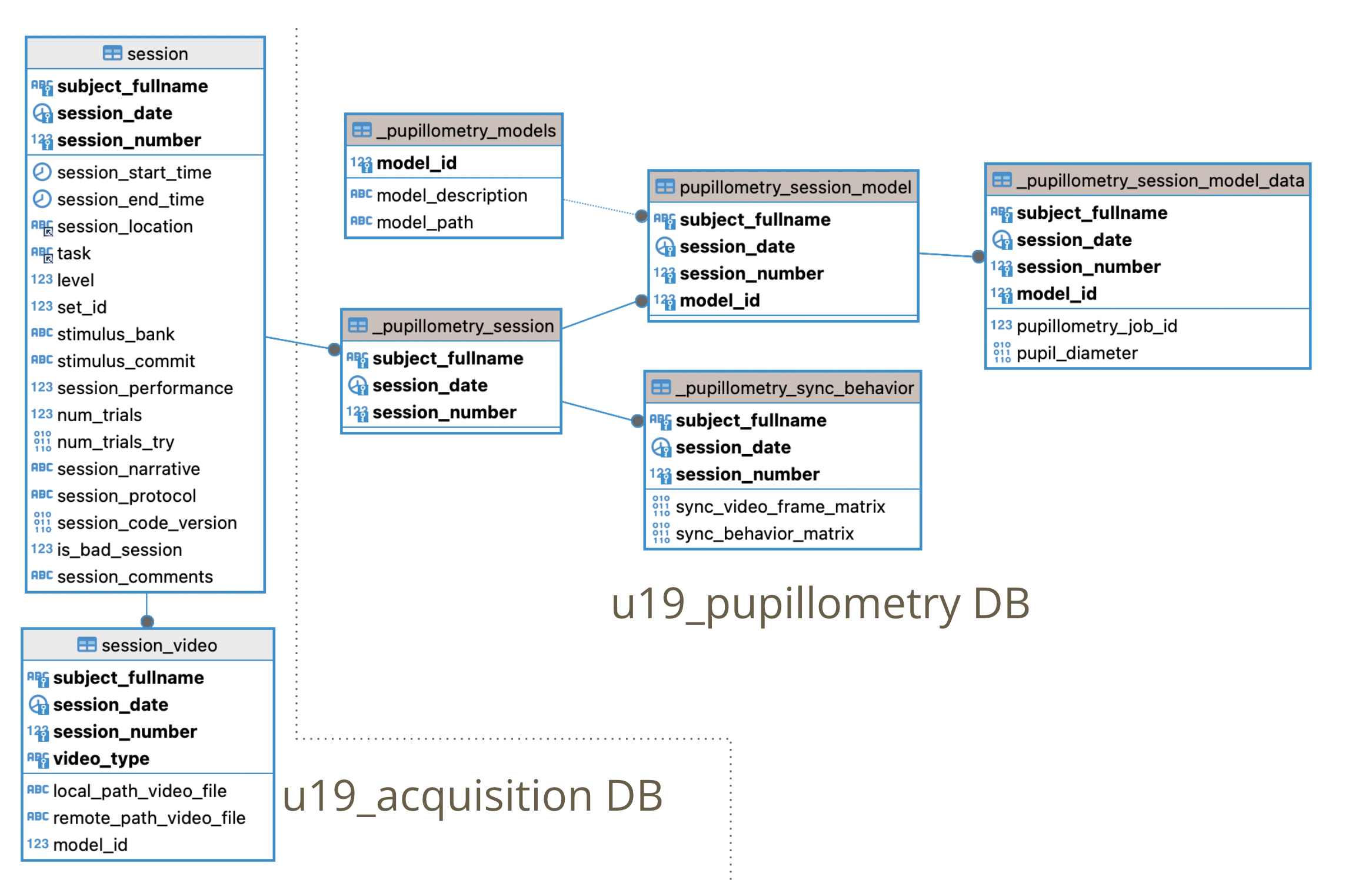
Steps executed in Pupillometry pipeline
When
pupillometry_video -> 2. In RigParametersis selected in training GUI a newsession_videorecord is registered in the DB while ingesting Session. (Code here)The
cmd_copy_video_filestask scheduler takessession_videorecords and store new videos in this location:\\cup.pni.princeton. edu\braininit\Data\Raw\video_pupillometry\(userid)\(subjectfullname)\(session_date)_g(session_number)\(video_file). Script run by pupillometry backup videos task scheduleDuring night cronjob: if video is found on cup location:
- Populate
pupillometry_sessiontable - Execute sync behavior code for video. Code here.
- Register default model to process video in
pupillometry_session_modelandpupillometry_session_model_data.
- Queue/Checks job in
spockvm2
Similar to ephys/imaging automation pipeline, pupillometry pipeline has a main (more simple) class to process all pupillometry jobs called
PupillometryProcessingHandler. This class is located here .Execute pupillometry_queue_jobs script. This scripts calls
PupillometryProcessingHandler.check_pupillometry_sessions_queuefunction. Queues a processing job to all sessions where a model was registered.Execute pupillometry_check_jobs script. This scripts calls
PupillometryProcessingHandler.check_processed_pupillometry_sessionsfunction. If processing job is finished get final result (pupil_diameter) and stores it inpupillometry_session_model_datatable.
Monitor pupillometry processing sessions
A few MATLAB functions were built to check how pupillometry jobs are doing:
Get processed data from a session:
key = struct('subject_fullname', 'efonseca_ef317_act116', 'session_date', '2024-02-21')
pupillometry_data = fetch(pupillometry.PupillometrySessionModelData * pupillometry.PupillometrySyncBehavior & key, '*')
pupillometry_data =
struct with fields:
subject_fullname: 'efonseca_ef317_act116'
session_date: '2024-02-21'
session_number: 0
model_id: 2
pupillometry_job_id: 2984
pupil_diameter: [108177×1 double]
sync_video_frame_matrix: [108177×4 single]
sync_behavior_matrix: [150227×5 single]
pupil_diameter: Pupil diameter for each video frame.
sync_video_frame_matrix: Time & Block/Trial/Iteration for each video frame.
sync_behavior_matrix: Time & video frame for each Block/Trial/Iteration.
- Restart processing for failed processing Sometimes processing fails for external factors (processing system was down, model was not properly selected, etc). As a first attempt to solve the issue a restart processing function was created:
psmd = pupillometry.PupillometrySessionModelData()
key = struct('subject_fullname', 'efonseca_ef317_act116', 'session_date', '2024-02-21')
psmd.restart_pupillometry_failed_job(key)
If processing fails again, contact Software Developer and check #automation_pipeline_errors slack channel for more information about the error.
- Check status for all sessions:
psmd = pupillometry.PupillometrySessionModelData()
all_sessions_table = psmd.check_status_pupillometry_jobs()
- Check status for a specific session(s):
psmd = pupillometry.PupillometrySessionModelData()
key = struct('subject_fullname', 'efonseca_ef317_act116', 'session_date', '2024-02-21')
session_status = psmd.check_status_pupillometry_jobs(key)
- Get all succesfully processed sessions:
psmd = pupillometry.PupillometrySessionModelData()
pupillometry_finished_sessions = psmd.get_finished_jobs_pupillometry()
Add a new video model to the pipeline
Check Deeplabcut documentation for step by step guide.
Ask for tips and tricks to our BRAINCoGS personnel that have created video models in the past:
- Joshua Julian (jjulian@princeton.edu)
- Juan Lopez (juanlopez@princeton.edu)
After video model has been created, copy the main directory of the model to this location:
\\cup.pni.princeton. edu\braininit\Data\Raw\video_models\(video_model_directory)Add model to the DB (u19_pupillometry.PupillometryModels table):
- MATLAB code:
new_model_key = struct()
new_model_key.model_description = 'New model to insert'
new_model_key.model_path = 'video_models/new_model_directory_name'
insert(pupillometry.PupillometryModels,new_model_key)
- Always add
video_models/to the model_path,before model_directory_name.
- Get
model_idof model you want to use for your sessions:
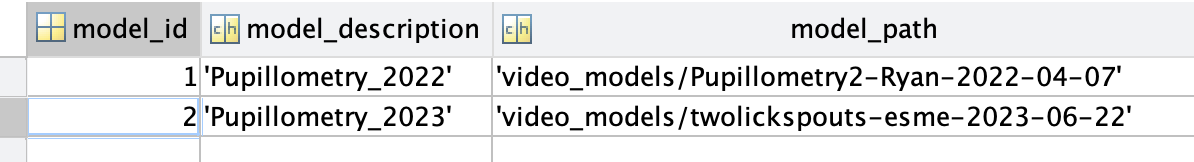
>> pupillometry.PupillometryModels
ans =
Object pupillometry.PupillometryModels
:: Table to store reference for each model ::
MODEL_ID model_description model_path
________ _____________________ ______________________________________________
1 {'Pupillometry_2022'} {'video_models/Pupillometry2-Ryan-2022-04-07'}
2 {'Pupillometry_2023'} {'video_models/twolickspouts-esme-2023-06-22'}
fetch(pupillometry.PupillometryModels,'*')
- Insert into
u19_pupillometry.PupillometrySessionModelsessions to be processed with the new model:
key = struct('subject_fullname', 'efonseca_ef317_act116', 'session_date', '2024-02-21')
key.model_id = 2
insert(pupillometry.PupillometrySessionModel, key, 'IGNORE');
- Sessions will be processed with new model overnight.