Ephys/Imaging Automation Pipeline in BRAINCoGS (Developer Guide)
The Ephys/Imaging Automation Pipeline in BRAINCoGS main goals are:
- Automate spike sorting and imaging segmentation for all recordings
- Centralize/Standardize paths for Recording Data Storage
- Unify & Register Ephys/Imaging Processing
- Store processed data in BRAINCoGS Database (DJ)
To accomplish this we developed three tools:
- Ephys/Imaging Automation GUI (RecordingProcessJobGUI)
- Recording Workflow management (Automatic_job directory in U19-pipeline_python )
- Collab reposiotries to handle Ephys/Imaging Processing (BrainCogsEphysSorters and BrainCogsImagingSegmentation )
Ephys/Imaging Automation GUI
In this mini guide for the automation GUI we will show the relationship between the GUI and the Database. From which tables some values are taken and which records are written by the GUI.
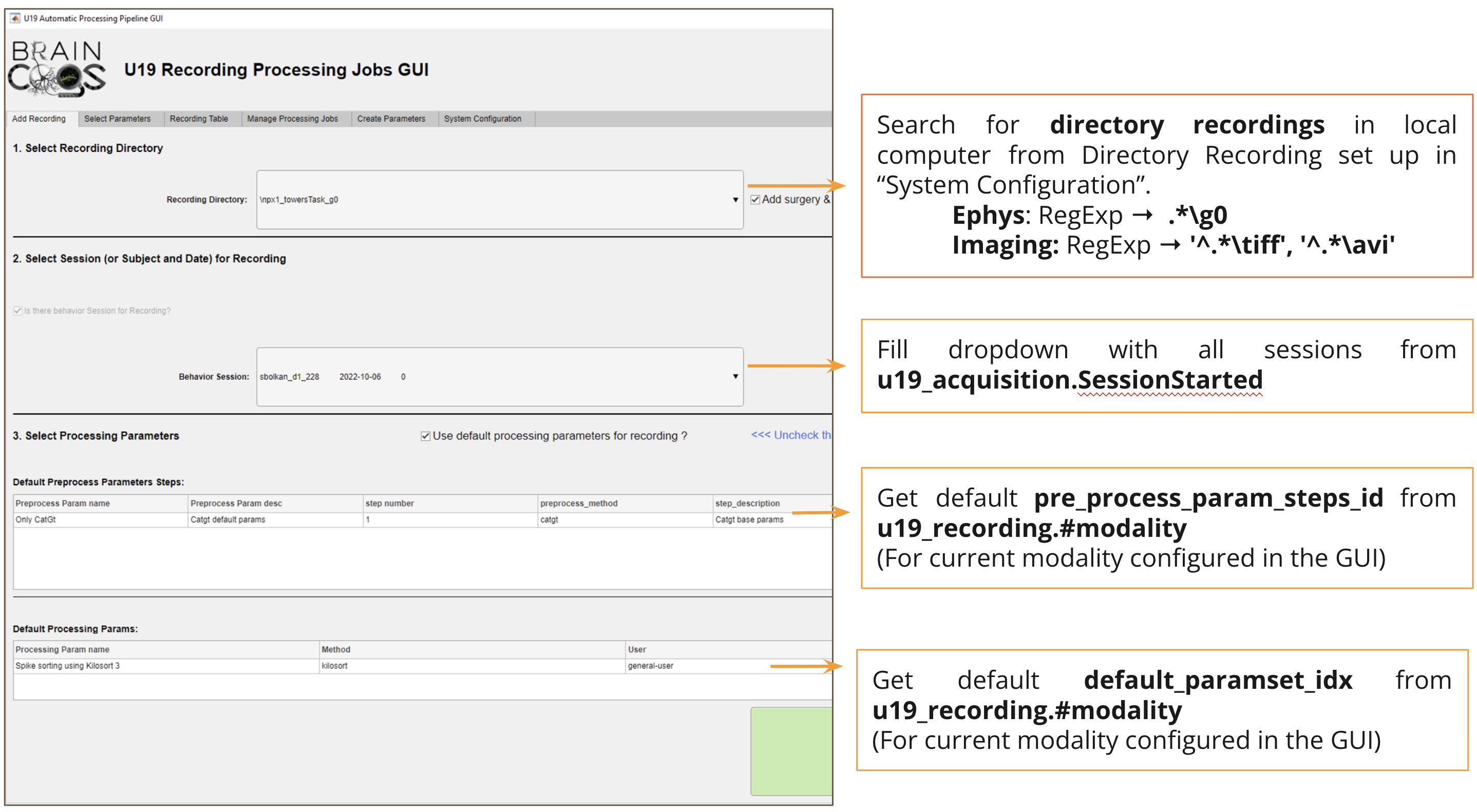
Automation GUI main screen
Ephys Preprocessing (precluster) parameters organization
Main tables
- u19_pipeline_ephys_element.#pre_cluster_method List of methods (or algorithms) supported for ephys preprocessing
- u19_pipeline_ephys_element.pre_cluster_param_set Specific set of parameters (mainly a dictionary) for a given preprocessing method. Multiple set of parameters can be stored for the same method.
- u19_pipeline_ephys_element.pre_cluster_param_steps (Ephys) Reference to a set to steps to perform in ephys preprocessing
- u19_pipeline_ephys_element.pre_cluster_param_steps__step These records indicate which set of parameters for given preprocessing methods will be executed (and in which order).
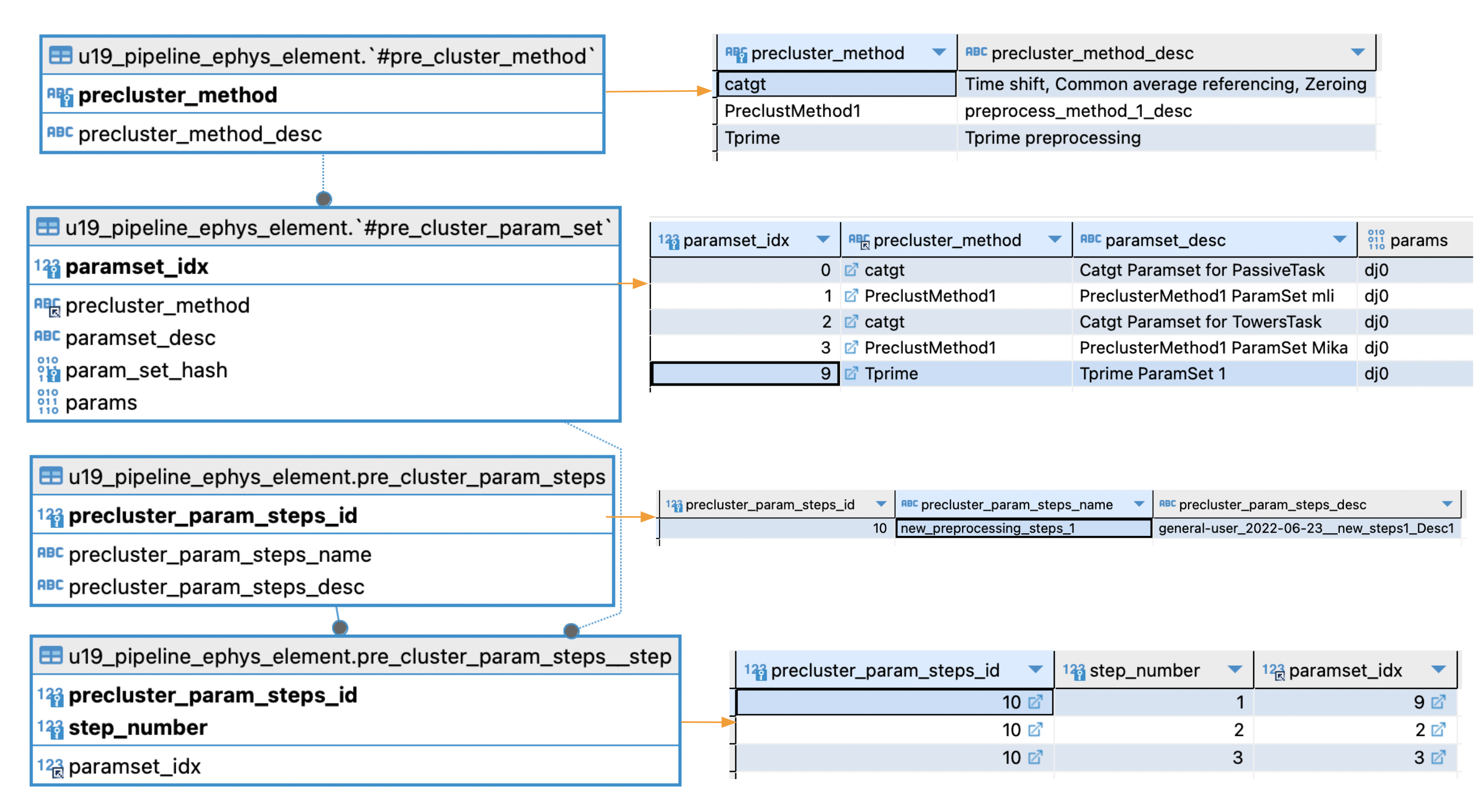
- Depicted in the above image:
- Let's pretend: precluster_param_steps_name = new_preprocessing_steps_1 (precluster_param_steps_id = 10 ) is selected for preprocessing.
- According to pre_cluster_param_steps__step
- paramset_idx = 9 will be executed 1st
- paramset_idx = 2 2nd
- paramset_idx = 3 3rd.
- Checking pre_cluster_param_set for paramset_idx = 9,2,3 we conclude preprocessing will comprise:
- Tprime (Tprime ParamSet 1)
- Catgt (Catgt ParamSet for Towers Task)
- PreClustMethod1 (PreClusterMethod1 Paramset Mika)
Epgys Processing (cluster) parameters organization
- Simpler than preprocessing structure (since there are no multiple steps involved), we have two tables to organize Ephys Processing parameters.
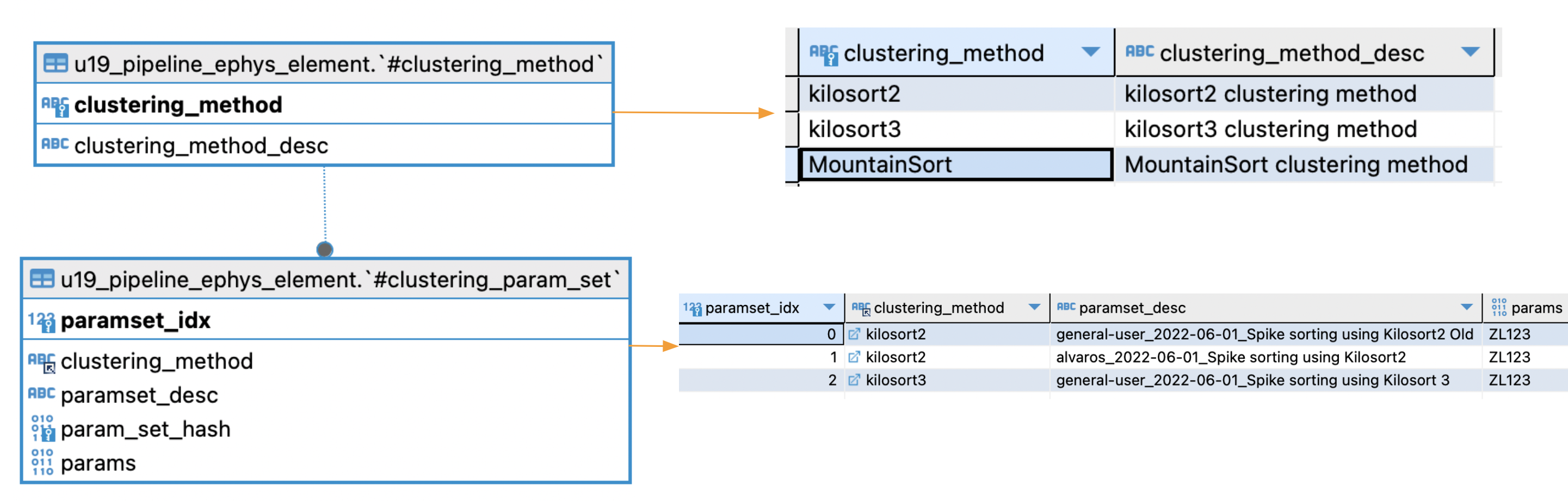
Main tables
u19_pipeline_ephys_element.#clustering_method List of methods (or algorithms) supported for ephys processing
u19_pipeline_ephys_element.#clustering_param_set Specific set of parameters (mainly a dictionary) for a given processing method. Multiple set of parameters can be stored for the same method.
Each recording (or to be precise, recording process) can be processed with a different set of parameters. Default parameters are used for the majority of the recordings in BRAINCoGS.
Default parameters for preprocessing and processing
- As seen in in Automation GUI main screen, u19_recording.#modality stores default parameters for each modality.
- As a developer manually update default parameters for all modalities when needed by the project.
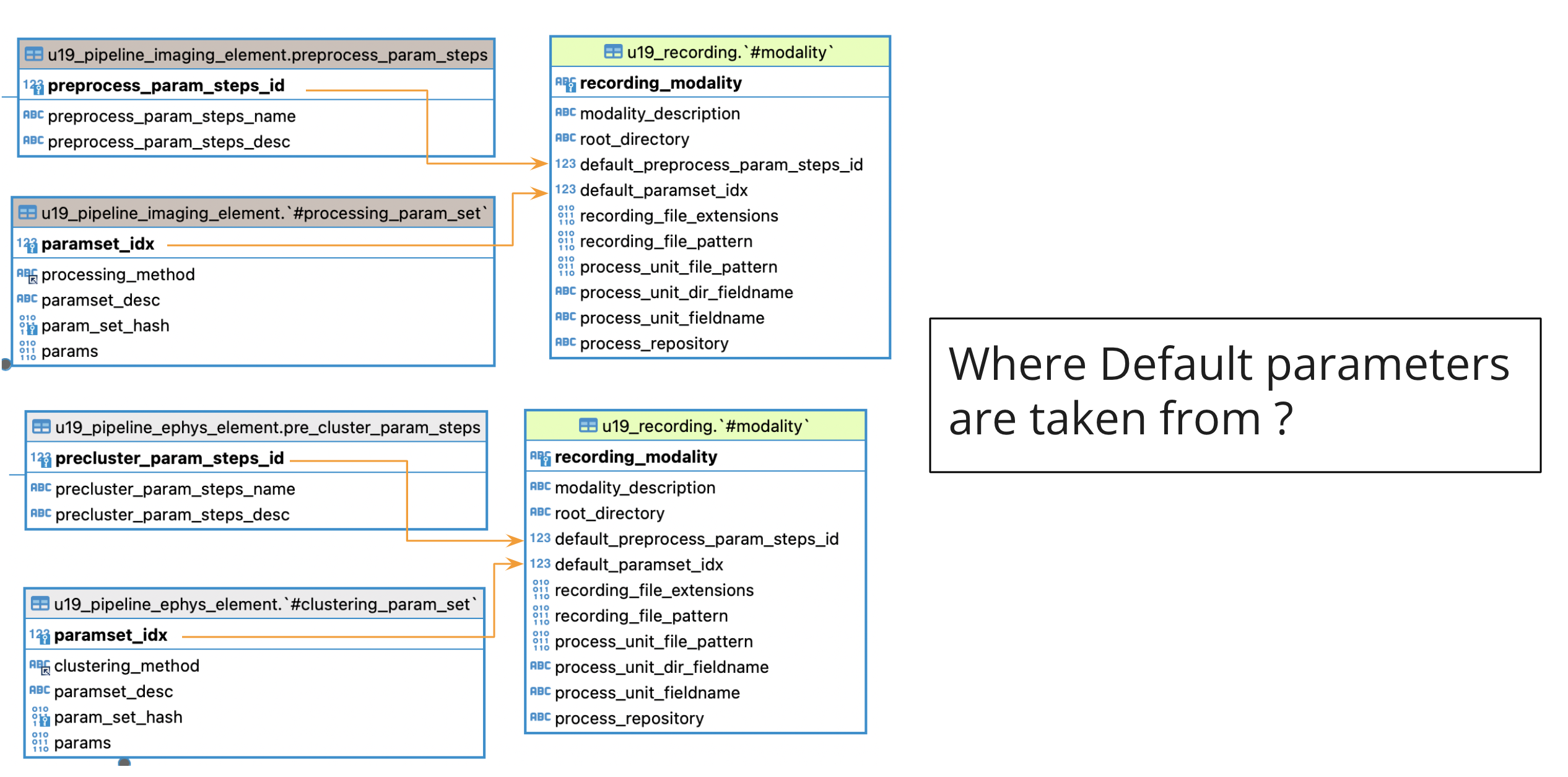
- In u19_recording.#modality table it is stored a reference for default parameters most commonly used for processing ephys & imaging.
- Main table to store preprocessing parameters:
- u19_pipeline_ephys_element.pre_cluster_param_steps: (Ephys) Reference to a set to steps to perform in ephys preprocessing
- Imaging, u19_pipeline_imaging_element.pre_process_param_steps: (Imaging) Reference to a set to steps to perform in imaging preprocessing (No preprocessing in imaging for any user at the moment)
- Main table to store processing parameters:
- Ephys, u19_pipeline_ephys_element.#clustering_param_set: (Ephys) Reference to a set of parameters for chosen sorting algorithm.
- Imaging, u19_pipeline_imaging_element.#processing_param_set: (Imaging) Reference to a set of parameters for chosen segmentation algorithm.
Imaging equivalence parameter tables:
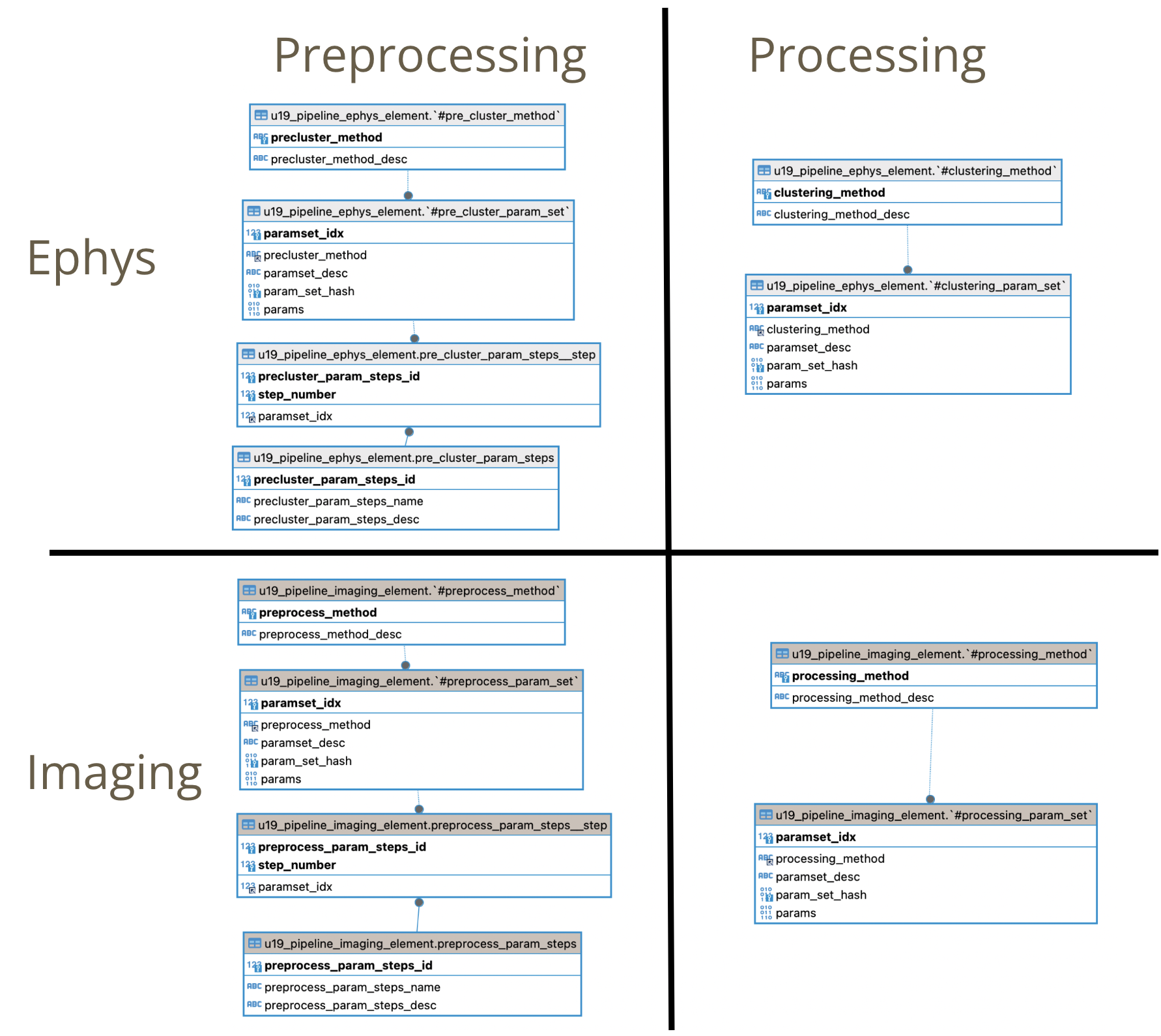
- All description made for ephys preprocessing and processing tables apply for the imaging counterparts.
Tables written when recording is registered:
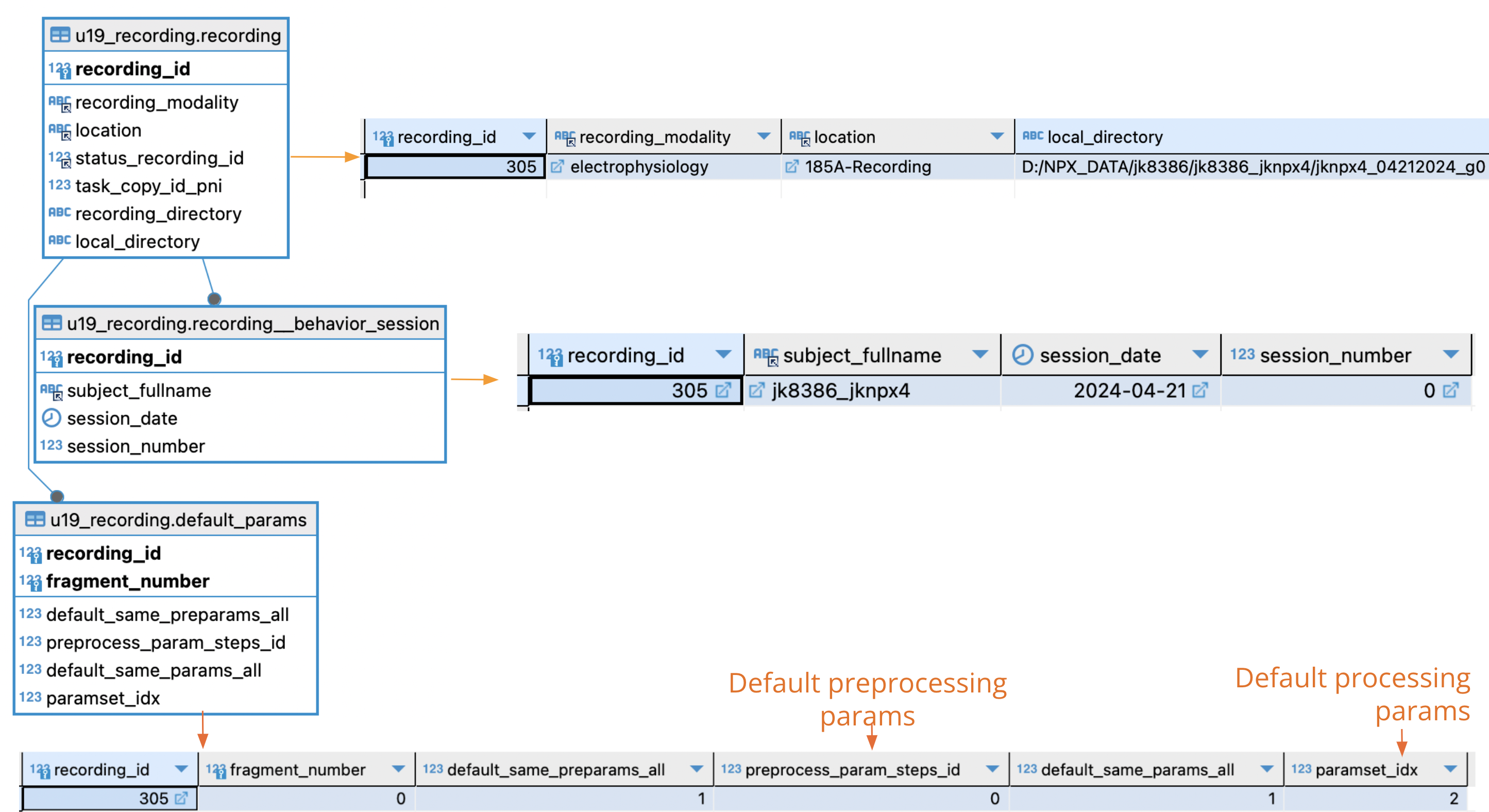
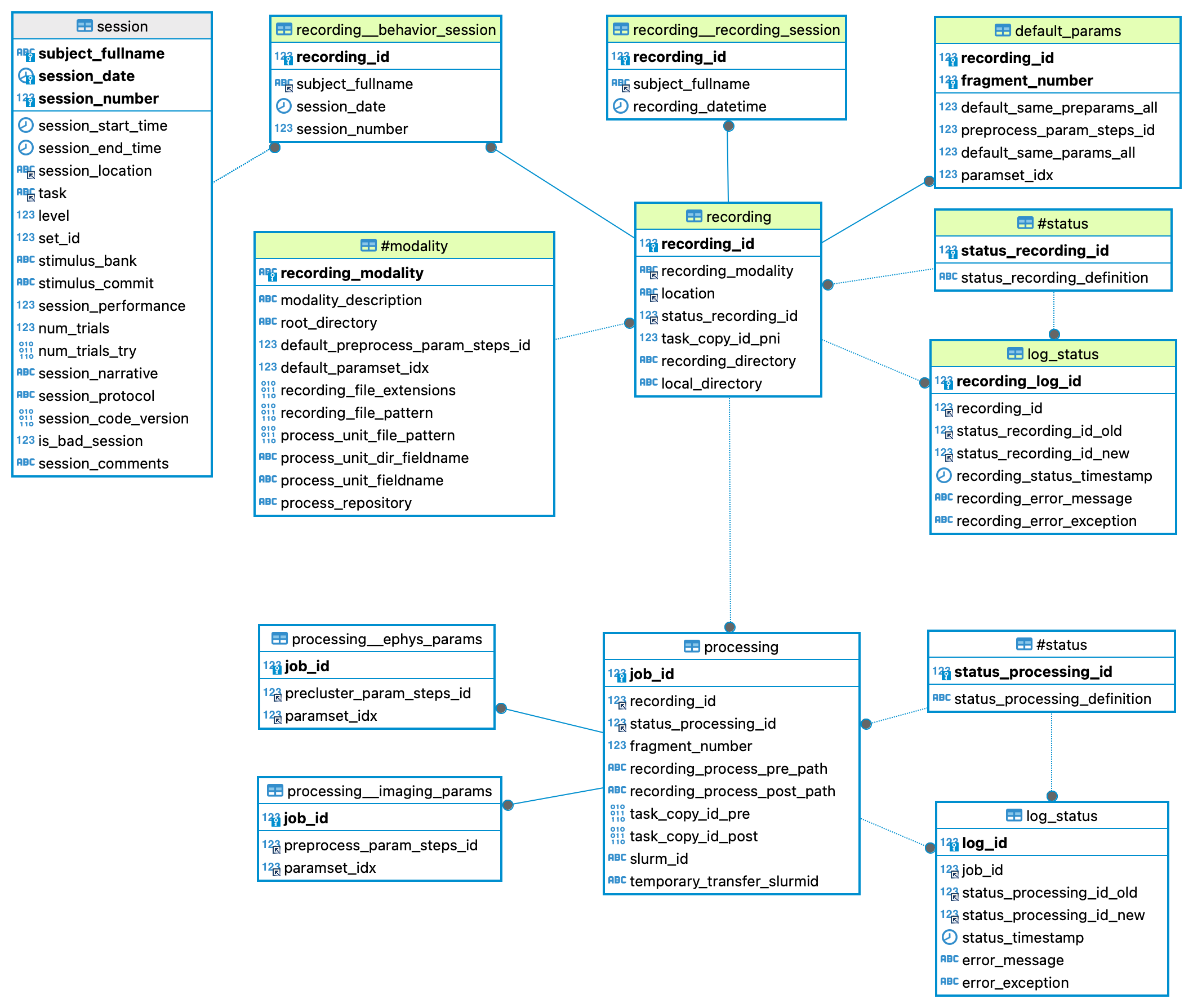
When a new recording is created three tables are written:
- u19_recording.recording: Main table for recordings. Recording_id is created will identify the recording through all the process
- u19_recording.recording__behavior_session: Reference to which behavior session corresponds this recording.
- u19_recording.default_params: Set of parameters chosen for this recording.
If there is no behavior session attached to recording:
- u19_recording.recording__recording_session: Subject and datetime of recording is stored as reference in this table.
u19_recording.default_params design:
- Default_params works as a "guide" to know which parameters where chosen for recording.
- Explanation for all fields of this table:
- recording_id Reference to which recording parameters are being selected
- fragment_number Reference to which "fragment" (or job) the parameters apply to. (Check next session to know how recordings are split in fragments).
- default_same_preparams_all If default_same_preparams_all = 1 (default case), same preprocessing parameters will be applied to all fragments of recording.
- preprocess_param_steps_id Preprocessing parameter id chosen for this recording-fragment. Taken from u19_recording.#modality by default.
- default_same_params_all If default_same_params_all = 1 (default case), same processing parameters will be applied to all fragments of recording.
- paramset_idx Processing parameter id chosen for this recording-fragment. Taken from u19_recording.#modality by default.
- In the default case (main screen Automation GUI) default_same_preparams_all=1 & default_same_params_all=1 so default parameters will be applied to all fragments of recording.
Workflow management description
Workflow management code creates and coordinates of a set of tasks for all recordings that were registered with the GUI to make sure they are entirely processed.
Shell code executed as a cronjob for workflow management: (call_cronjob_automatic_job.sh )
Workflow management is composed mainly by two classes that handles recordings and recording_processes (recording_processes or jobs are how recordings are composed)
- Ephys recordings are composed by one or many independent probe electrophysiology recordings. Each probe recording correspond to a job in the workflow management
- Calcium imaging recordings are composed by one or many independent field of views image stacks. Each field of view image stack correspond to a job in the workflow management.
The class that manages workflow at the recording level is (RecordingHandler)
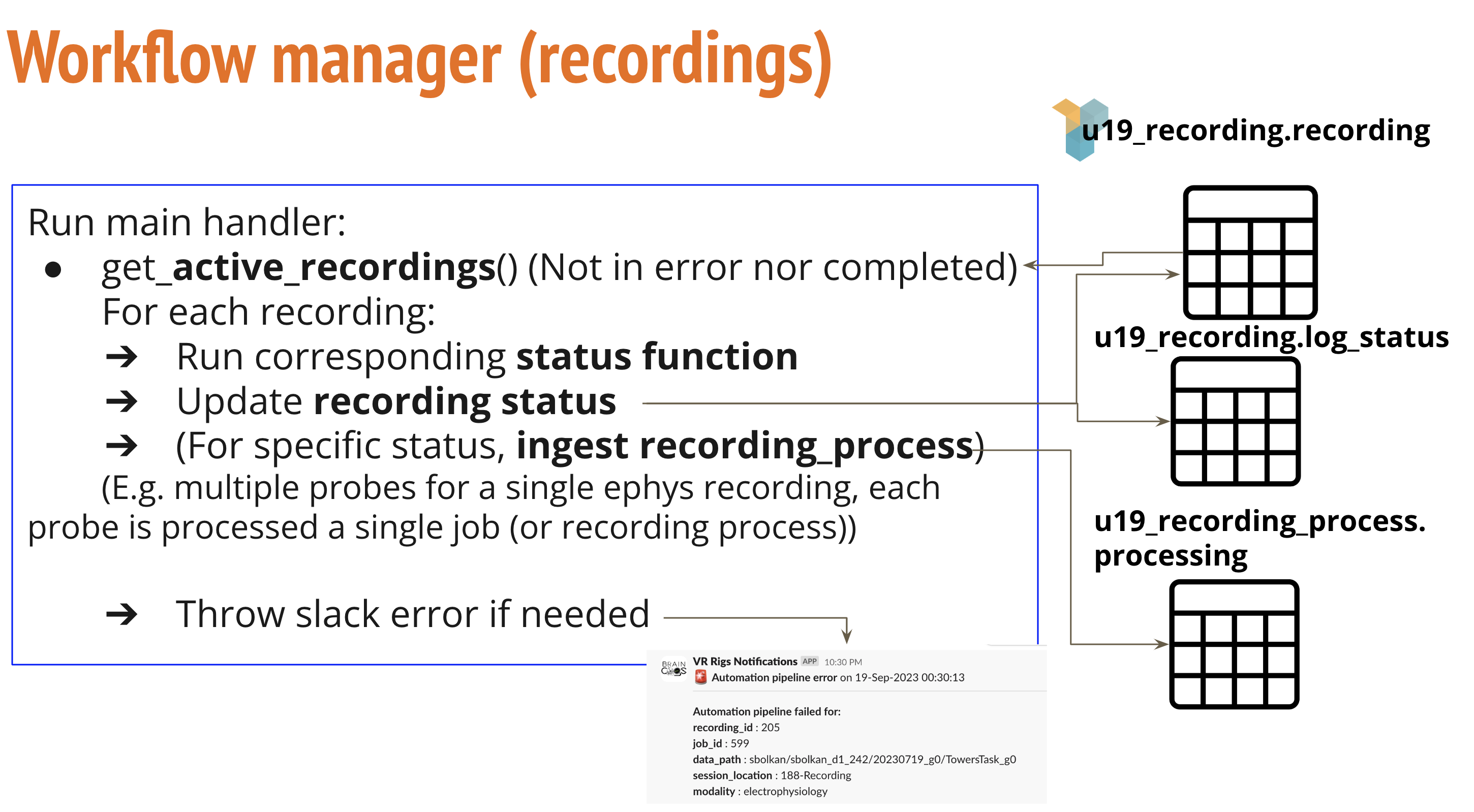
Main functions and variables in recording workflow manager
- recording_status_dict in (Params Config file): This dictionary defines status definitions and corresponding functions to execute.
- pipeline_handler_main in (RecordingHandler): Main function in recording workflow
- Executes corresponding functions based in status.
- Executed every 30 minutes to check for new recordings to be handled.
- Send notifications for processed and failed functions.
- exception_handler in (RecordingHandler): Python decorator for error handling.
- modality_preingestion in (RecordingHandler): Main ingestion function from recording to recording_process tables. There are subcalls depending on modality of recording (ephys or imaging).
Imaging preingestion main steps:
- imaging_preingestion in (RecordingHandler): Ingestion to recording_process table for an imaging recording. Get all FOVs (TIFF stacks) for the recording and assign a new job for each one with corresponding parameters fetched from selection done in automation GUI. AcquiredTiff populate function in (Imaging pipeline): Auxiliar script to call populate_Imaging_AcquiredTiff script in MATLAB.
- populate_Imaging_AcquiredTiff in (populate_Imaging_AcquiredTiff): Population calls to:
- u19_imaging_pipeline.AcquiredTiff: Each recording is divided into Tiff Splits (e.g. Mesoscope recordings contain multiple tiff stacks that will be processed independently). (Code here)
- u19_imaging_pipeline.SyncImagingBehavior: Find correspondence between virtual reality frame in the behavior experiment and Calcium Imaging frame in recording. (Code here)
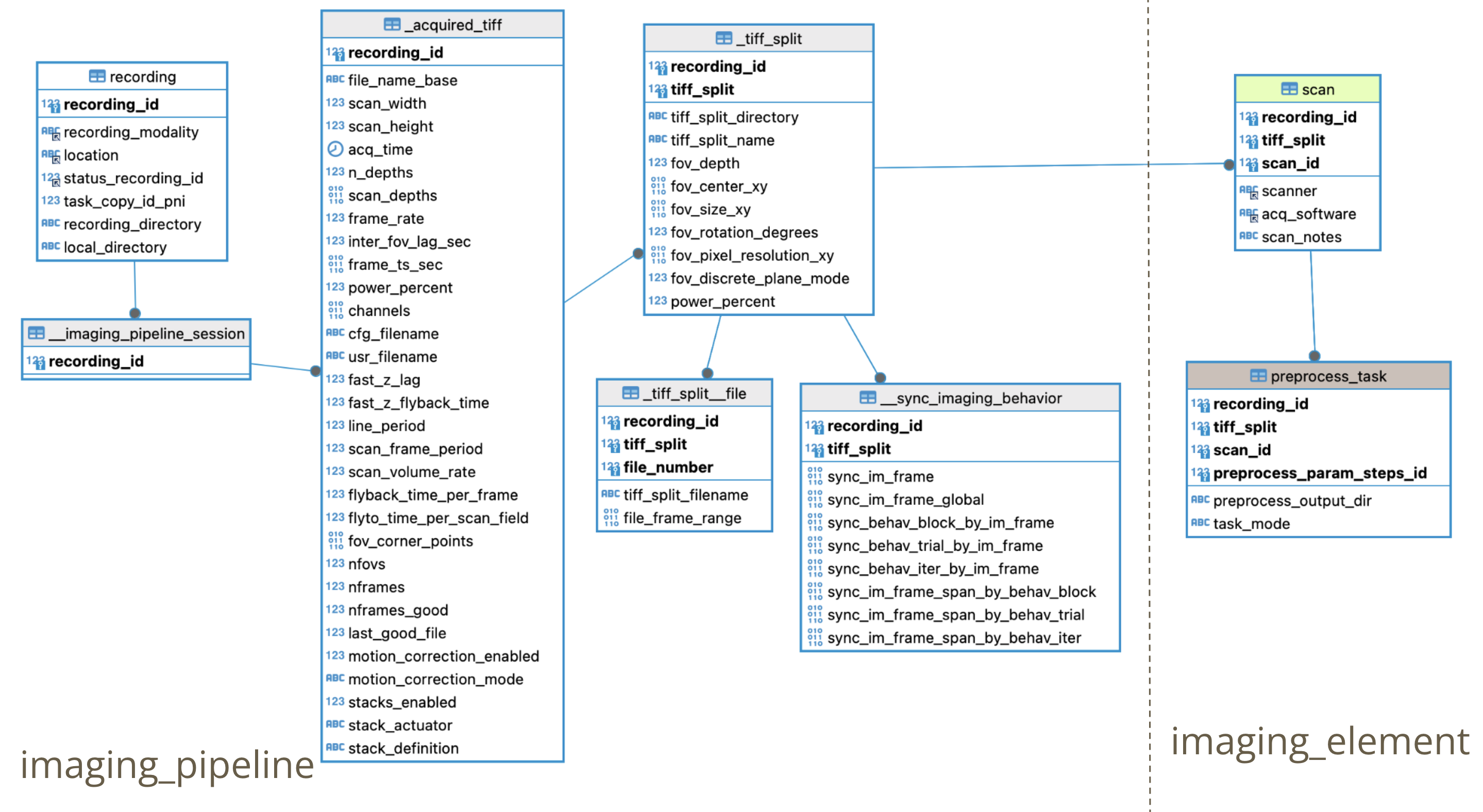
Ephys preingestion main steps:
- electrophysiology_preingestion in (RecordingHandler): Ingestion to recording_process table for an ephys recording. Get all probes for the recording and assign a new job for each one with corresponding parameters fetched from selection done in automation GUI.
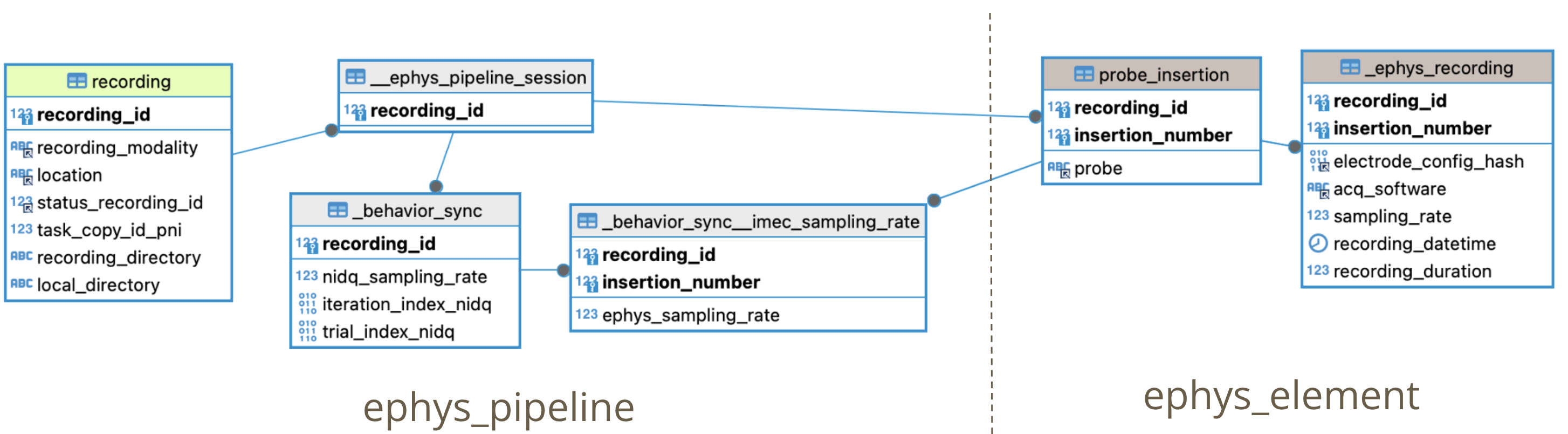
- Ingest ephys_pipeline.EphysPipelineSession table
- Ingest ephys_element.ProbeInsertion table
- Ingest ephys_element.EphysRecording table
- Ingest ephys_pipeline.BehaviorSync table: Find corresponding iteration in ephys recording with frame from Virmen behavior task (Code here ) ( and here ).
- For each probe (insertion_number) in EphysSession insert a Processing (job) in u19_recording_process.Processing
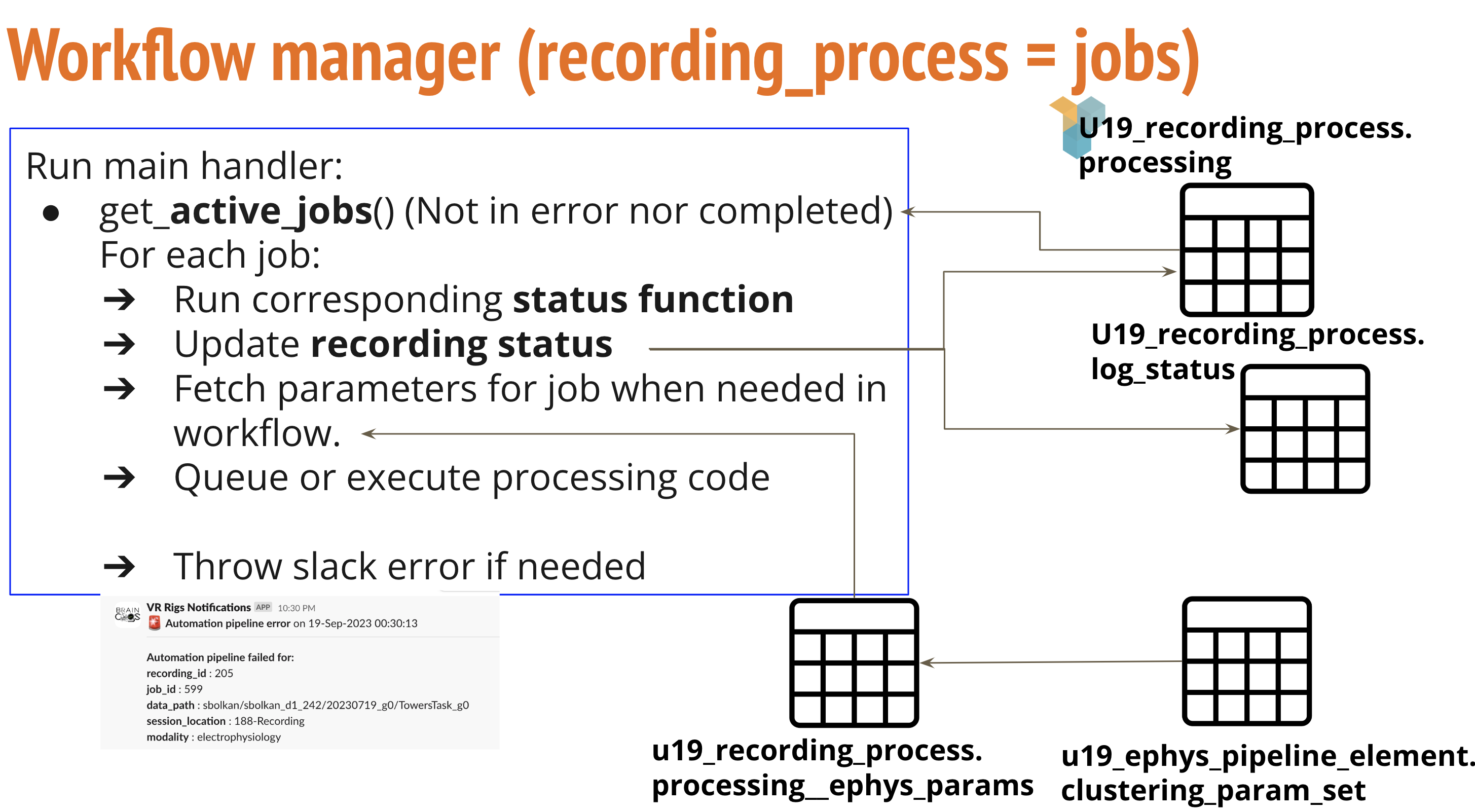
Main functions and variables in recording_process workflow manager
- recording_process_status_dict in (Params Config file): This dictionary defines status definitions and corresponding functions to execute.
- pipeline_handler_main in (RecProcessHandler): Main function in recording process workflow
- Executes corresponding functions based in status.
- Executed every 30 minutes to check for new recordings to be handled.
- Send notifications for processed and failed functions.
- transfer check/review in (transfer_check/review): Executes and monitors globus transfer from PNI to PrincetonUniversity clusters. (Deprecated)
- slurm_job_queue/check in (slurm_job_functions): Generate slurm file and queue the job in the cluster that will process recording process. Monitor job to check if it has already finished.
- populate_element in (slurm_job_queuew): After processing jobs populate imaging or ephys element tables downstream from results file.
Collab reposiotries to handle Ephys/Imaging Processing
BrainCogsEphysSorters
Set up instructions for BrainCogsEphysSorters in cluster system
Set up new processing cluster
Instructions to set up a new computing cluster to process Ephys & Imaging