Manipulation pipeline
- This documentation will guide the researcher through the process of creating a new manipulation pipeline.
- At BRAINCoGS optogenetics and thermal manipulation are currently supported.
What does the “manipulation” pipeline include:
- Minimum data framework to store in a DB all relevant data from a specific manipulation.
- Behavior integration. Training system will include the manipulation as an option to be selected for a behavior session.
- Generic software parameters to be used in behavior code.
Prerequisites
- In order to create a new manipulation it’s assumed that:
- The researcher is able to connect to datajoint00.pni.princeton.edu DB
- Latest version of u19_pipeline_matlab repository.
Initial set-up
- Connect to database
connect_datajoint00 - Create new manipulation schema (substitute manipulation_name with the real name of the manipulation:
create_new_manipulation_schema('(manipulation_name)', 1) - This will create a new schema “base” code on the
U19-pipeline-matlab/schemasdirectory: - (We will use “thermal” manipulation for this example).

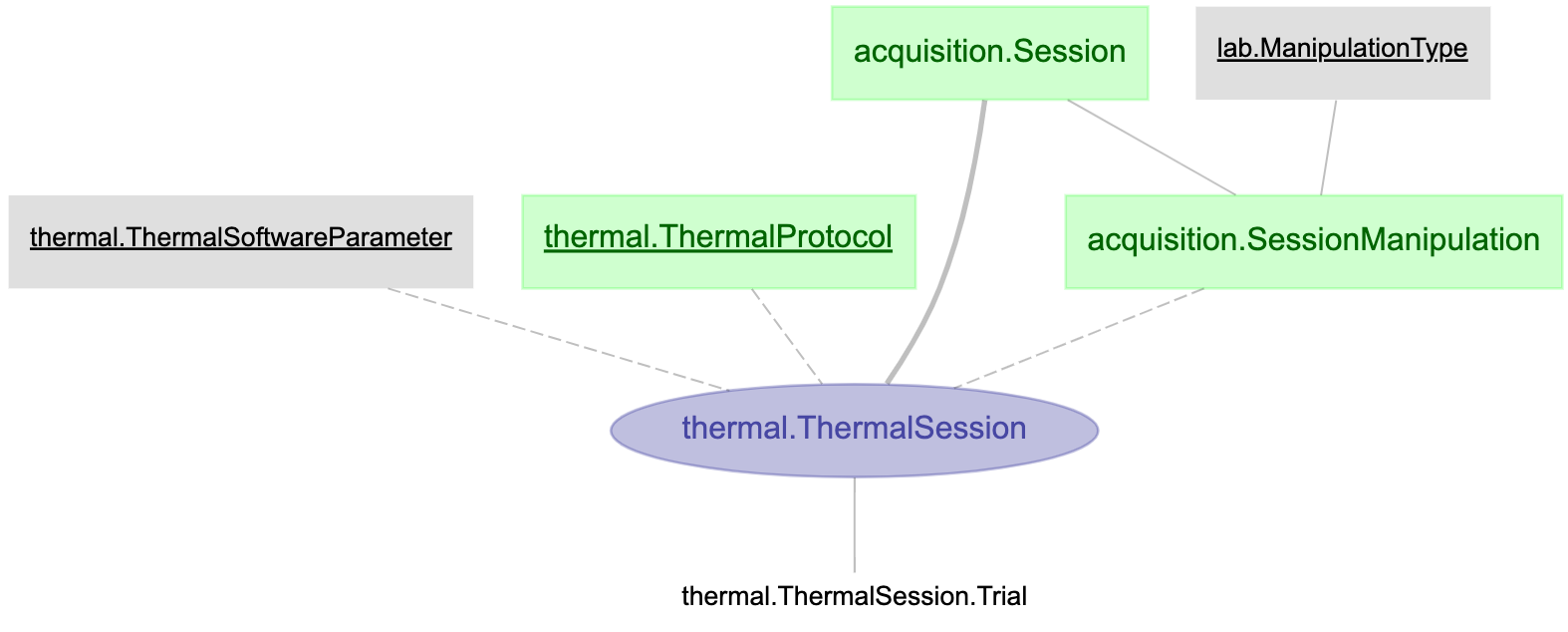
Table description
- Throughout the table description chapter we are going to give an example of an already working manipulation pipeline. (Optogenetics)
"Manipulation" Protocol table
The protocol table stores related information that defines the current manipulation “type” to be used on a behavior session.
Here is the minimum table definition for a manipulation protocol table, it is composed by an id to identify the protocol and a description field.
Generic "Manipulation" Protocol.m
%{
# Defined <manipulation> protocols for training
<manipulation>_protocol_id : int AUTO_INCREMENT
---
protocol_description : varchar(256)
%}
Adding features to "Manipulation" Protocol table
For each manipulation protocol it is possible to add from 0 to n “features” that will define & describe the protocol. We are going to describe all features added for OptogeneticsProtocol as an example:
It is important to know from an optogenetic experiment what kind of stimulation was given to the subject: Frequency, wavelength, power etc. All these variables can be stored into a “feature” table and be categorized as StimulationParameters.
What if stimulation was not a square pulse ? We can create a “feature” table to define (if needed) specific waveforms for a given session. (OptogeneticsWaveform)
What if different rooms have different laser systems models ? We can create a “feature” table to store all possible devices to be used in an optogenetic experiment (OptogeneticsDevice).
For each of these features we need to create a new table that encompasses the needed information for that feature. We will call all these extra tables a protocol “feature” table.
For a guide on how to define DJ tables go to: this link.
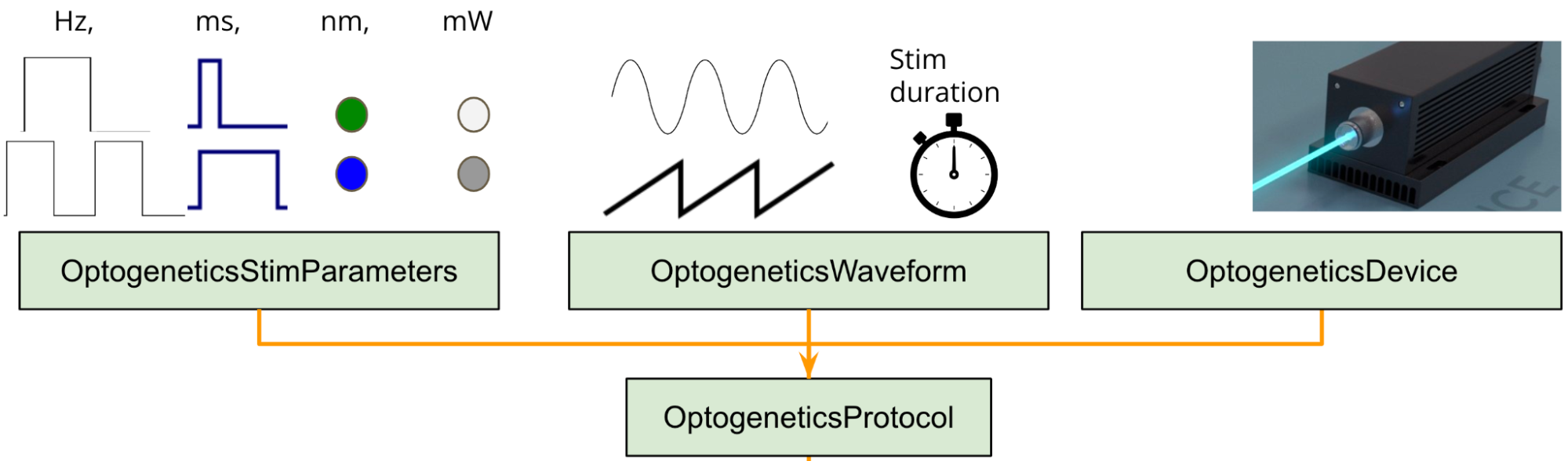
- For the current guide we will only show OptogeneticsStimulationParameters definition as an example:
OptogeneticsStimulationParameters.m:
%{
# Parameters related to laser stimulation
stim_parameter_set_id : int AUTO_INCREMENT #
---
stim_parameter_description : varchar(256) #
stim_wavelength : decimal(5,1) # (nm)
stim_power : decimal(4,1) # (mW)
stim_frequency : decimal(6,2) # (Hz)
stim_pulse_width : decimal(5,1) # (ms)
%}
classdef OptogeneticStimulationParameter < dj.Lookup
properties
end
end
Fields needed for a protocol “feature” table:
id field: as an int AUTO_INCREMENT type as the only primary key (e.g. stim_parameter_set_id).
extra_fields: Any other field that helps to define the feature.
After all feature tables are defined they should be added to the "Manipulation" Protocol table.
For our Optogenetics example:
% Declare new "feature" table
optogenetics.OptogeneticsStimulationParameters
% Add the feature -> protocol table
add_feature_key_protocol_table(optogenetics.OptogeneticsProtocol, ... optogenetics.OptogeneticsStimulationParameters)
% Sync definition from DB to .m file
syncDef(optogenetics.OptogeneticsProtocol);
% clear previous connection and connect again
clear all
connect_datajoint00
- After the “features” tables are added to the "Manipulation" Protocol table we are ready to add protocols to be “ready” and selectable for a behavior session:
% Insert stim parameter record
stim_parameter_rec.stim_parameter_description = 'cool stims'
stim_parameter_rec.stim_wavelength = 473
stim_parameter_rec.stim_power = 10
stim_parameter_rec.stim_frequency = 100
stim_parameter_rec.stim_pulse_width = 1
insert(optogenetics.OptogeneticsStimulationParameters, stim_parameter_rec)
% get last inserted stim_id
stim_id = fetch(optogenetics.OptogeneticsStimulationParameters, 'ORDER BY stim_parameter_set_id desc LIMIT 1');
% or look for a previously inserted parameter
all_stim_params = fetch(optogenetics.OptogeneticsStimulationParameters, '*')
stim_id = 1;
% Insert new protocol with new stimulation parameter
new_protocol.protocol_description = 'this_is_new_protocol'
new_protocol.stim_parameter_set_id = stim_id;
insert(optogenetics.OptogeneticsProtocol, new_protocol)
"Manipulation" SoftwareParameters table
- The software parameters table stores a set of parameters (a matlab struct, a python dictionary) that the code that handles the behavior will use during the session.
- We will show how to insert new software parameters:
- This for the optogenetics.OptogeneticSoftwareParameter table
param_struct = struct();
param_struct.software_parameter_description = 'stimulation_sequence # 1';
% All parameters goes in here
%(P_on and lsrepoch are the common and needed for current opto experiments)
param_struct.software_parameters.P_on = 0.21;
param_struct.software_parameters.lsrepoch = 'cue';
%Insert parameter
software_param_id = try_insert(optogenetics.OptogeneticSoftwareParameter, param_struct)
- Check insert_optogenetic_software_parameter script to use as example.
- How to read software parameters on experiment code (ViRMEn)
- Example to get software parameters on the initializatonCodeFun on virmen:
function vr = initializationCodeFun(vr)
vr.software_params = vr.exper.userdata.trainee.softwareParams.software_parameters;
vr.lsrepoch = vr.software_params.lsrepoch;
vr.P_on = vr.software_params.P_on;
"Manipulation" Session table
- This table stores manipulation data for a specific behavior session. This table “links” a manipulationProtocol & manipulationSoftwareParameters with a behavior Session.
- This table does not need any additional code on it. (Unless extra fields from the behavior file are needed to be stored). Researcher should contact DB designer if that is their intention
OptogeneticSession.m
%{
# Information of a optogenetic session
-> acquisition.Session
---
-> acquisition.SessionManipulation
-> optogenetics.OptogeneticProtocol
-> optogenetics.OptogeneticSoftwareParameter
%}
"Manipulation" SessionTrial table
- This table stores data, on a trial by trial basis, corresponding to the manipulation performed during the behavior session.
- There is a section on any "Manipulation" SessionTrial class on the get_manipulation_trial_data function code where researcher has to add lines to fetch specific trial manipulation data:
Code extract from OptogeneticSessionTrial table
function trial_structure = get_manipulation_trials_data(~,session_key, log)
.
.
for itrial = 1:nTrials
curr_trial = log.block(iBlock).trial(itrial);
trial_data = session_key;
trial_data.stim_on = curr_trial.lsrON;
trial_data.t_stim_on = time_trial(curr_trial.iLaserOn);
trial_data.stim_epoch = num2str(curr_trial.LaserTrialType);
trial_structure(total_trials) = trial_data;
Training with new manipulation
- After all code for new manipulation has been set up the researcher will be able to select a specific manipulation type, protocol & software parameters that will be associated with the schedule for a given animal. Subsequent behavior sessions will correspond to that selection.
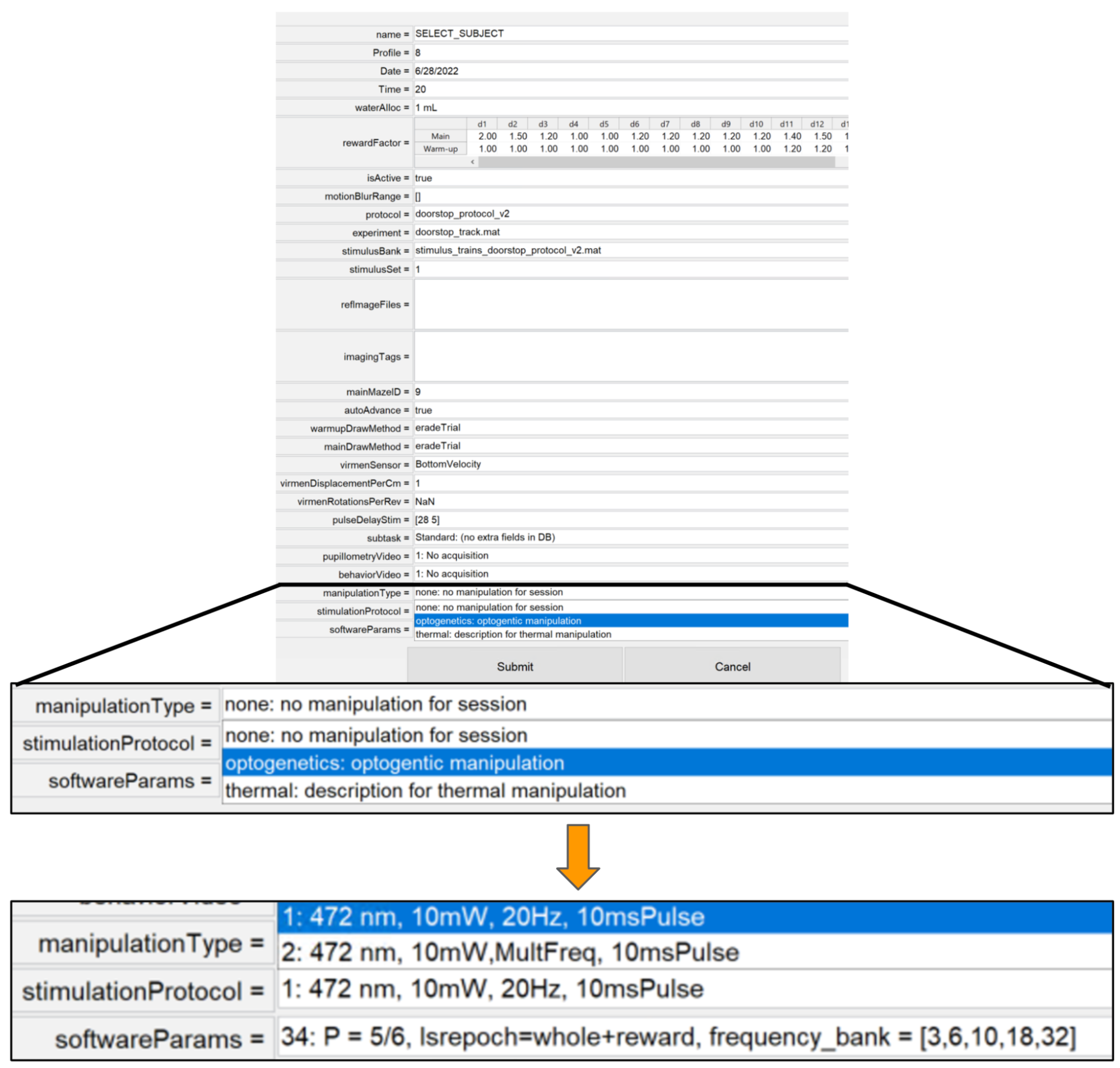
Fetching Data
- After training has occurred all relevant data will be accessible in the corresponding tables of the database.
- Datajoint fetch guide
key = struct('subject_fullname', 'sbolkan_a2a_492', 'session_date', '2022-06-27')
fetch(optogenetics.OptogeneticSessionTrial * optogenetics.OptogeneticSession & key,'*')
ans =
363×1 struct array with fields:
subject_fullname
session_date
session_number
block
trial_idx
stim_on
t_stim_on
t_stim_off
stim_epoch